Beef
marbling measurement using spectral imaging: A multiple linear regression
approach
Medición del
marmoleado de carne de res usando imágenes espectrales: Un enfoque de regresión
lineal múltiple
Victor Aredo1,
*; Lía Velásquez2; Nikol Siche3
1 Departamento de Operaciones
Unitarias. Facultad de Química e Ingeniería Química, Universidad Nacional Mayor
de San Marcos, Av. República de Venezuela s/n, Lima, Peru. varedot@unmsm.edu.pe
2 Department of
Food Engineering, Faculty of Animal Science and Food Engineering, University of
São Paulo, Pirassununga, SP 13635-900, Brazil. lia.vc@usp.br
3 Escuela de Ingeniería
Zootecnista, Facultad de Ciencias Agropecuarias, Universidad Nacional de
Trujillo, Av. Juan Pablo II s/n, Trujillo, Peru. nsichei@unitru.edu.pe
*
Autor corresponsal: varedot@unmsm.edu.pe (V. Aredo).
ID
ORCID de los autores
V.
Aredo:  https://orcid.org/0000-0002-3293-7696 L.
Velásquez:
https://orcid.org/0000-0002-3293-7696 L.
Velásquez:  https://orcid.org/0000-0001-8091-5611
https://orcid.org/0000-0001-8091-5611
N.
Siche:  https://orcid.org/0000-0002-7174-8337
https://orcid.org/0000-0002-7174-8337
ABSTRACT
This study aimed at
measuring beef marbling scores in an objective and simple manner through spectral
imaging and multiple linear regression (MLR). Beef marbling prediction by
hyperspectral imaging and partial least squares regression (PLSR) was analyzed to
calibrate and evaluate an MLR model with a few selected wavelengths. Data came
from 44 beef samples and consisted of their spectral signatures (75
wavelengths) from hyperspectral reflectance images (400-1000 nm) and their
marbling scores assigned by evaluators. The wavelengths that presented
regression coefficients with the highest absolute values in the PLSR model,
were used to calibrate the MLR model by a backward stepwise approach (p <
0.05). The coefficient of determination for prediction (R2p)
and the standard error of prediction (SEP) were evaluated. The MLR model was suitable
for practical use because it required only 12 wavelengths for reliable
predictions (R2p = 0.824 > 0.8; SEP = 11.4% < 15%).
A model is proposed for the objective and simple measurement of beef marbling
score using multispectral imaging technology.
Keywords: hyperspectral
imaging; partial least squares regression; predictive model; meat quality;
multispectral imaging; multiple linear regression.
RESUMEN
Este estudio tuvo como objetivo medir el
puntaje de marmoleado de carne de vacuno de una manera objetiva y sencilla
mediante imágenes espectrales y regresión lineal múltiple (RLM). La predicción
del marmoleado de la carne usando imágenes hiperespectrales y regresión de
mínimos cuadrados parciales (RMCP), fue analizada para para calibrar y evaluar un
modelo RLM con algunas longitudes de onda seleccionadas. Los datos provinieron
de 44 muestras de carne de vacuno y consistieron en sus firmas espectrales (75
longitudes de onda) de imágenes de reflectancia hiperespectral (400-1000 nm) y las
puntuaciones de marmoleado asignadas por los evaluadores. Las longitudes de
onda que presentaron coeficientes de regresión con valores absolutos más altos
en el modelo RMCP se utilizaron para calibrar el modelo RLM mediante un enfoque
por pasos hacia atrás (p < 0,05). Se evaluaron el coeficiente de
determinación de predicción (R2p) y el error estándar de
predicción (ESP). El modelo RLM era adecuado para uso práctico porque requería
sólo 12 longitudes de onda para predicciones confiables (R2p =
0,824 > 0,8; ESP = 11,4% < 15%). Se propone un modelo para la medición
objetiva y sencilla del puntaje de marmoleado de carne de vacuno mediante
tecnología de imágenes multiespectrales.
Palabras
clave:
imágenes
hiperespectrales; regresión parcial de mínimos cuadrados; modelo predictivo;
calidad de la carne; imágenes multiespectrales; regresión lineal múltiple.
Recibido: 24-09-2023.
Aceptado: 02-11-2023
Beef is one of the most consumed foods of animal
origin. It is a source of high-biological value proteins and minerals, such as
iron and zinc, vitamin A and B-complex vitamins (Liu et al., 2022; Vidal et
al., 2022; Pereira & Vicente, 2013). Nowadays, beef production seeks to
satisfy the demanding quality requirements of consumers. One of the quality
attributes of interest to consumers is marbling, which consists of the amount
and spatial distribution of intramuscular fat in the longissimus dorsi muscle
(Gagaoua et al., 2022; Khaled et al., 2021). The consumers perceive this
attribute as white specks or streaks in the lean muscle. Highly marbled steaks
and other cuts of beef provide higher palatability (i.e., better sensory
quality), which results in a higher marbling score (Khaled et al., 2021; Cheng
et al., 2015).
Beef producers estimate or measure marbling as a
reference to establish the commercial value of their product. The beef marbling
is commonly measured manually using meat graders, which can be affected by the
fatigue and natural subjectivity of the evaluator. In this context, several
efforts have focused on developing marbling measurement methods using modern
technologies, such as infrared spectroscopy, Raman spectroscopy, color
computational imaging, and multi/hyperspectral imaging (Pinto et al., 2023;
Echegaray et al., 2022; Cheng et al., 2015). Among them, multi/ hyperspectral
imaging stands out since it combines digital imaging and spectroscopy,
providing spatial and chemical spectral information about the samples (Jia et
al., 2022; Özdoğan et al., 2021; Siche et al., 2016).
The high potential of multi/hyperspectral imaging to
measure marbling and other quality attributes in the beef industry 4.0, is due
to the application of data science strategies that handle the collinear and
high-dimensional data to propose solutions with only the necessary data (Nychas
et al., 2021; Echegaray et al., 2022). For measuring the beef marbling using
multi/hyperspectral imaging, one strategy was to binarize sample images with a
decision tree to determine fat area and subsequently apply a quadratic
regression model to calculate beef marbling scores (Velásquez et al., 2017).
Another strategy was to binarize images based on the maximum (fat/lean) ratio
of gray value to determine the area of large, medium, and small fat particles
and subsequently calculate meat marbling scores using a multiple linear
regression (MLR) model (Li et al., 2011). In our previous study (Aredo et al.,
2017), a partial least squares regression (PLSR) model was proposed to
calculate beef marbling from the spectral signatures of the samples;
considering procedural aspects, it can be affirmed that this model used a
simple approach.
The solution to predictive tasks based on a calibrated
PLSR model with full spectra can be simplified by calibrating an MLR model with
selected spectral data (ElMasry et al., 2007). MLR models stand out for being
easy to calibrate and understand for end users, which facilitates their
practical application in industry through multi-spectral imaging technology (Su
& Sun, 2018). In this sense, the present study explored measuring beef
marbling scores by spectral imaging and MLR with a few selected wavelengths.
Data
acquisition
This work used data from
Aredo et al. (2017), where beef marbling score was predicted using
hyperspectral imaging. Procedures and equipment for data acquisition and
treatment, are briefly described here. A total of 58 beef samples (longissimus
dorsi muscle) from Chachapoyas (Amazonas, Peru) were collected. These samples
were divided into two sets: (1) a set of 44 samples for model calibration and
(2) a set of 14 samples for prediction using the model.
The samples were scanned in
a hyperspectral imaging system with a broom approach in reflectance mode. The
hyperspectral imaging camera was a Pica XC model (Resonon Inc., USA), and the
image acquisition/processing software was SpectrononPRO (Resonon Inc., USA).
The system captured hyperspectral images in a spectral range of 400-1000 nm
with a spectral resolution of 8 nm (i.e., 75 wavelengths were explored). The
spectral signatures consisted of the average spectra in the region of interest
from the hyperspectral images of each sample.
The reference measurement
of the marbling score of the samples was made through the visual appraisal of
15 evaluators. It consisted of assigning scores by comparing photographs of the
samples with the beef marbling standard of the Japan Meat Grading Association
(JMGA, 2000), which ranges from 1 (minimum marbling) to 12 (maximum marbling).
Regression
The marbling
scores of the samples assigned by evaluators were the dependent variable, while
the spectral signature (75 wavelengths) of the samples was the dependent
variable.
The calibration
of the PLSR model by Aredo et al. (2017) for the prediction of beef marbling
score using hyperspectral imaging was reviewed. PLSR is a machine learning
technique introduced by Geladi & Kowalski
(1986) used in chemometric tasks (Wold et al., 2001). It explains the
descriptors by means of orthogonal factors, also called latent variables. The
optimal number of latent variables is identified with the minimum mean squared
prediction error value. Subsequently, the regression coefficients for the
optimal number of latent variables are obtained.
Regression coefficients of
PLSR models calibrated with near-ideal numbers of latent variables were also
generated. This step was carried out with the intention of facilitating the
identification of the wavelengths that carry important information for the
prediction through the criterion of the highest local values of the regression
coefficients.
The spectral signature of
the samples was reduced considering the selected wavelengths. Using the reduced
spectral signature and the marbling value assigned by
the sample evaluators, a model was calibrated using multiple linear regression
(MLR). This regression method is the most used when there is a linear
relationship between the variables and is based on the principle of minimizing
the prediction error (Çetin et al., 2022). In this work, this regression was
used with the aim of a potential practical implementation of the simplified
model through multispectral imaging technologies (Xie et al., 2018) and to
facilitate intuitive recalibration for users with little mathematical / statistical
/ computational knowledge. For comparative purposes, a reduced PLSR model was
calibrated with the same data used for the MLR model.
Calibration of
the PLSR model was performed using Matlab 2023a software (MathWorks, Natick,
MA, USA), while calibration of the MLR model was performed using MS Excel
software (Microsoft INC). Both PLSR and MLR models, being linear models, can be
represented by eq. (1) or by eq. (2) in detail.
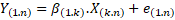 (1)
(1)
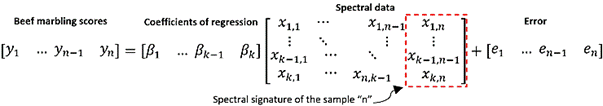 (2)
(2)
In the case of
the PLSR model, n: is the number of samples (44), k: is the number of
wavelengths in the spectral signature (75), Y: is the response matrix (1 x 44)
of marbling score values, β: is the matrix
of regression coefficients (1 x 75), X: is the matrix of predictive variables
constituted by the spectral signatures of the samples (75 x 44) and e: is the
matrix of residual information not explained by the model (1 x 44).
In the case of
the MLR and reduced PLSR models, n: is the number of samples (44), k: is the
number of selected wavelengths identified from the analysis of the regression
coefficients of the PLSR model, Y: is the response matrix (1 x 44) of marbling
scores values, β: is the matrix of regression coefficients (1 x k), X: is
the matrix of predictive variables constituted by the reduced spectral
signatures (selected wavelengths) of the samples (k x 44) and e: is the matrix
of unexplained residual information by template (1x44).
The performance
of the models was evaluated in the calibration set using the coefficient of
determination of calibration (R2c) and standard error of
calibration (SEC), and in the prediction set using the coefficient of
determination of prediction (R2p) and standard error of
prediction (SEP) (ElMasry et al. 2007). The SEC and SEP are represented in eqs.
(3-5).
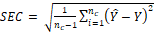 (3)
(3)
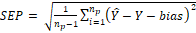 (4)
(4)
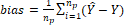 (5)
(5)
where, Ŷ:
is the predicted marbling score values; Y: the measured marbling score values;
nc: the number of samples (spectral signatures) in the calibration
set (44); and np: number of samples (spectral signatures) in the
prediction set (14) and the “bias”: is the systematic error.
The figure 1
outlines the mentioned steps.
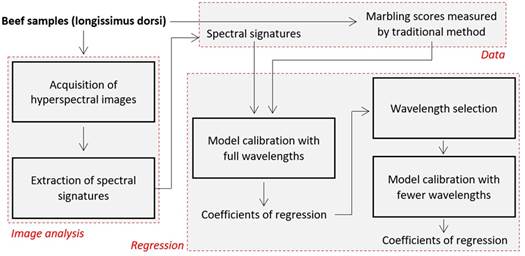
Figure 1. Main steps
for measurement of beef marbling score by spectral imaging.
The steps of this study are the basic ones for the predictive
tasks followed in the literature. There are studies where some complementary analyses
are carried out for a better understanding or handling of the data.
2.3 Complementary analyses
A principal component analysis was performed on the spectral data
from the calibration set. This technique reduces the dimension of the data and
extracts the variance of the data by projecting it into principal components
(Bro & Smilde, 2014). This study focused on interpretating scores and
loadings on the principal components to get insights about the samples and
wavelengths, respectively (Khan et al., 2020). The scores plot was used to
observe the beef marbling scores of the samples according to the spectral data.
The loadings plot was used to identify the selected wavelengths based on the
spectral data.
The coefficient of correlation between each wavelength and beef
marbling scores was determined. It was carried out to identify the wavelengths
that can potentially be used to predict the quality attribute of interest (Dong
et al., 2016).
These
analyses were performed using Matlab 2023a software (MathWorks, Natick, MA,
USA).
Full model verification
Verification of the calibration of the PLSR model
by cross-validation (Figure 2) revealed that the ideal number of latent
variables was 15, and that a similar number of latent variables (13, 14 and 16)
can produce models with a low error level.
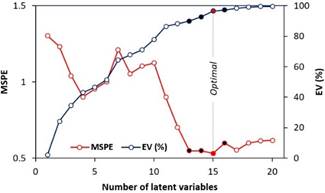
Figure 2. Mean Squared
Prediction Error (MSPE) and Explained Variance (EV) in Partial Least Squares
Regression (PLSR) models with different number of latent variables.
The analysis of regression coefficients of the
PLSR model is commonly performed in the one calibrated with the optimal number
of latent variables (15). However, as the PLSR models calibrated with 13, 14
and 16 latent variables present a low error, it was considered interesting to
analyze the coefficients of these PLSR models together (Figure 3).
The remarkable overlapping of the regression
coefficients provides evidence that the wavelengths that carry essential information
for predicting marbling are similar to each other over the range of interest of
the latent variables. Therefore, wavelengths with regression coefficients with
higher absolute values should be useful for calibrating simplified models.
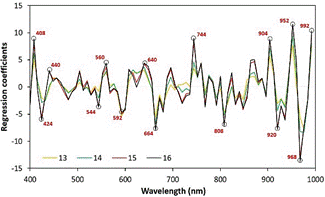
Figure 3. Coefficients
of Partial Least Squares Regression (PLSR) models calibrated with 13-16 latent
variables.
MLR calibration
The calibration of the MLR model (Table 1) using a stepwise
procedure with a backward elimination approach revealed that the selected
wavelengths that had the highest absolute values in the PLSR regression
coefficient were significant (p < 0.003). The MLR
model was statistically valid (p = 0.00)
for predicting beef marbling (Table 1), and it has a predictive performance
lower than the full PLSR model (Table 2), which is expected when less spectral
information is used (from 75 to 12 wavelengths). On the other hand, the
calibration of a PLSR model with the same reduced data and 12 latent variables
yielded similar results to the MLR model (Table 2). Considering the simplicity,
the MLR model is suitable for practical application because it achieves
acceptable predictive performance (R2p > 0.8, SEP <
15%) with reduced data (Siche et al., 2016; Su & Sun, 2018). In addition,
the visual verification of the performance of the MLR model (Figure 4)
indicates that it is adequate because both the calibration and prediction set
points are close to the ideal line.
Table
1
Regression
coefficients of the multiple linear regression model
|
Wavelength
(nm)
|
Coefficients
|
Error
|
t-statistical
|
Significance
|
|
Interception
|
-3.898
|
1.711
|
-2.28
|
0.030
|
|
408
|
0.788
|
0.226
|
3.49
|
0.001
|
|
560
|
4.833
|
1.393
|
3.47
|
0.002
|
|
592
|
-9.330
|
1.710
|
-5.46
|
0.000
|
|
640
|
13.514
|
2.739
|
4.93
|
0.000
|
|
664
|
-12.589
|
2.428
|
-5.19
|
0.000
|
|
744
|
7.691
|
1.524
|
5.05
|
0.000
|
|
808
|
-6.475
|
1.166
|
-5.55
|
0.000
|
|
904
|
16.752
|
3.855
|
4.35
|
0.000
|
|
920
|
-14.094
|
4.143
|
-3.40
|
0.002
|
|
952
|
15.374
|
3.426
|
4.49
|
0.000
|
|
968
|
-22.576
|
3.508
|
-6.43
|
0.000
|
|
992
|
6.388
|
1.856
|
3.44
|
0.002
|
Analysis of variance: FCal=21.56
> FTab = 2.08, p = 0.00.
Table 2
Performance of models in
predicting beef marbling score
|
Type of model
|
Number of
wavelenghts
|
Calibration
|
|
Prediction
|
|
R2c
|
SEC
|
|
R2p
|
SEP
|
SEP (%)
|
|
Full PLSR*
|
75
|
0.962
|
0.2
|
|
0.908
|
0.3
|
6.6
|
|
MLR
|
12
|
0.893
|
0.4
|
|
0.824
|
0.5
|
11.4
|
|
Simplified
PLSR
|
12
|
0.891
|
0.4
|
|
0.819
|
0.5
|
11.6
|
PLSR: Partial Least Squares
Regression, MLR: Multiple Linear Regression, R2c: coefficient of determination for calibration, SEC: standard
error of calibration, R2p: coefficient of determination of prediction, SEP: standard
error of prediction.
* Aredo et al
(2017).
A point to highlight is the limitation of the
model, as it is calibrated with samples with marbling scores between 1.2 and 5.3
(Figure 4); it cannot be used for reliable predictions in samples with marbling
greater than 5.3. However, this does not rule out the possibility that the
selected wavelengths (Table 1) are used as a reference in the recalibration of
MLR models for multispectral systems with beef samples with different marbling without
the need for extensive statistical knowledge of the user.
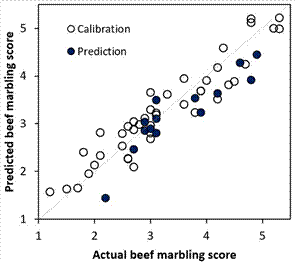
Figure 4. Actual vs.
predicted beef marbling score using the reduced multiple linear regression
model.
Regarding the selected
wavelengths for beef marbling score measurement, it is worth mentioning that
some of them are similar to those used in the evaluation of the composition of
beef by near-infrared spectroscopy. It is well known that the visible region of
the spectrum (400-700 nm) is related to the pigments in the samples. In this
sense, pigments, such as oxymyoglobin and myoglobin, usually have strong
absorption in this region (Alomar et al., 2003). Some reported wavelengths are
the 430 nm wavelength called the Soret absorption band in the blue region of
the spectrum due to the heme protein, which is attributed to trace amounts of
hemoglobin in meat; and the 574 nm wavelength that is related to oxyhemoglobin
absorption (Cozzolino & Murray, 2002). In near-infrared region, the
wavelengths of 908 and 928 nm are attributed to proteins and fats, respectively
(Alomar et al., 2003). Furthermore, the 970 nm wavelength is related to the
second O-H stretching overtone due to the presence of water in the sample
(Cozzolino & Murray, 2002; Alomar et al., 2003).
Complementary analyses
Principal component
analysis revealed that 99.31% of the variance of the spectral data was captured
with the first and second principal components (Figure 5 and Figure 6). In the
score plot (Figure 5), it is not possible to differentiate samples with
different beef marbling scores. It indicates that the high variance captured
from the spectral data by the principal components does not necessarily imply
class differentiation due to the unsupervised nature of
the technique (Munera et al., 2018).
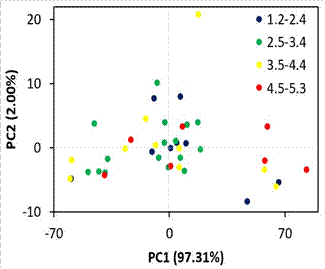
Figure 5. Scores plot
(principal component 1 vs principal component 2) of spectral data of samples with
different beef marbling scores.
The loadings plot (Figure 6) revealed
that the loadings of the wavelengths are positive in the first principal
component, while part of the loadings are negative in the second principal
component. The loadings appear to form a continuous “curve” with peaks,
troughs, and inflection points. These elements could be used as a reference to suggest
the selected wavelengths to predict the quality attribute of interest. However,
in this case, with only this result, it is difficult to choose these
wavelengths precisely.
The loadings plot including the
wavelengths selected by multiple linear regression analysis (Figure 6),
confirms that most of the selected wavelengths are in positions that define the
“curve”. An interesting fact is that the selected wavelengths are “balanced” concerning
the principal component 2; that is, six of them have positive values, and the
other six have negative values. Moreover, the selected wavelengths have
relatively high values in principal component 1, except for the 408 nm
wavelength. This suggests that the selected wavelengths for predicting quality
attributes can be confirmed with the loadings plot of principal component
analysis.
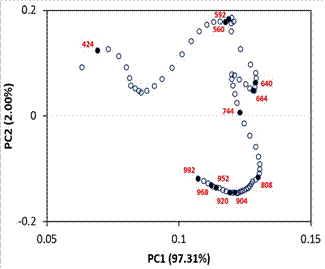
Figure 6. Loadings plot
(principal component 1 vs principal component 2) of spectral data of beef
samples with different marbling scores*.
* Black points are the
loadings of wavelengths used in multiple linear regression model.
The evaluation of the
correlation coefficient of each wavelength with actual beef marbling scores
(Figure 7), revealed that all the wavelengths were weakly correlated with beef
marbling scores. In this sense, this result cannot support the selection of
wavelengths to calibrate simplified linear models (Aredo et al., 2019).
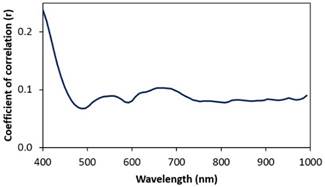 Figure
7. Correlation coefficients (r) between each spectral wavelength
with actual beef marbling scores.
Figure
7. Correlation coefficients (r) between each spectral wavelength
with actual beef marbling scores.
This study
demonstrated that is possible to measure beef marbling using spectral imaging
and MLR with acceptable predictive performance (R2p = 0.824
> 0.8, SEP = 11.4% < 15%). The MLR used only 12 wavelengths, which is
suitable for practical application by multispectral imaging technology. This
model is simpler and easy to understand; hence, it has the potential to help
beef producers with a low understanding of data science to obtain information
on the quality of their product for decision-making regarding market pricing.
The model features also facilitate recalibration tasks in other realities.
V.
Aredo thanks the USP ESALQ MBA Scholarship Program - USP Community. N. Siche
thanks the Universidad Nacional de Trujillo for the financial support to carry
out student mobility at the Universidad Nacional de Colombia, Medellín campus
(RR N° 1393-2023-UNT).
Alomar,
D., Gallo, C., Castaneda, M., & Fuchslocher, R. (2003). Chemical and
discriminant analysis of bovine meat by near infrared reflectance spectroscopy
(NIRS). Meat
science,
63(4), 441-450. https://doi.org/10.1016/S0309-1740(02)00101-8
Aredo,
V., Velásquez, L., & Siche, R. (2017). Prediction of beef marbling
using hyperspectral imaging (HSI) and partial least squares regression (RMCP). Scientia
Agropecuaria,
8(2), 169-174. http://dx.doi.org/10.17268/sci.agropecu.2017.02.09
Aredo,
V., Velásquez, L., Carranza-Cabrera, J., & Siche, R. (2019). Predicting of
the Quality Attributes of Orange Fruit Using Hyperspectral Images. Journal
of food quality and hazards control, 6, 82-92.
http://dx.doi.org/10.18502/jfqhc.6.3.1381
Çetin,
N., Karaman, K., Kavuncuoğlu, E., Yıldırım, B., &
Jahanbakhshi, A. (2022). Using hyperspectral imaging technology and machine
learning algorithms for assessing internal quality parameters of apple fruits. Chemometrics
and Intelligent Laboratory Systems, 230, 104650.
https://doi.org/10.1016/j.chemolab.2022.104650
Bro,
R., & Smilde, A. K. (2014). Principal component analysis. Analytical methods,
6(9), 2812-2831. https://doi.org/10.1039/C3AY41907J
Cheng,
W., Cheng, J. H., Sun, D. W., & Pu, H. (2015). Marbling analysis for
evaluating meat quality: Methods and techniques. Comprehensive Reviews in
Food Science and Food Safety, 14(5), 523-535.
https://doi.org/10.1111/1541-4337.12149
Cozzolino,
D., & Murray, I. (2002). Effect of sample presentation and animal muscle
species on the analysis of meat by near infrared reflectance spectroscopy. Journal
of Near Infrared Spectroscopy, 10(1), 37-44.
Dong,
J., Guo, W., Wang, Z., Liu, D., & Zhao, F. (2016). Nondestructive
determination of soluble solids content of ‘Fuji’apples produced in different
areas and bagged with different materials during ripening. Food Analytical
Methods, 9, 1087-1095. https://doi.org/10.1007/s12161-015-0278-4
Echegaray,
N., Hassoun, A., Jagtap, S., Tetteh-Caesar, M., Kumar, M., Tomasevic, I., ... ,
& Lorenzo, J. M. (2022). Meat 4.0: Principles and applications of Industry
4.0 technologies in the meat industry. Applied Sciences, 12(14), 6986.
https://doi.org/10.3390/app12146986
ElMasry,
G., Wang, N., ElSayed, A., & Ngadi, M. (2007). Hyperspectral imaging for
nondestructive determination of some quality attributes for strawberry. Journal
of food engineering, 81(1), 98-107.
https://doi.org/10.1016/j.jfoodeng.2006.10.016
Gagaoua, M., Duffy, G., Álvarez García, C., Burgess, C., Hamill,
R., Crofton, E. C., ... & Troy, D. (2022).
Current research and emerging tools to improve fresh red meat quality. Irish Journal of Agricultural and Food Research, 61(1), 145-167. https://doi.org/10.15212/ijafr-2020-0141
Geladi,
P., & Kowalski, B. R. (1986). Partial least-squares regression: a tutorial.
Analytica chimica acta, 185, 1-17. https://doi.org/10.1016/0003-2670(86)80028-9
Jia,
W., van Ruth, S., Scollan, N., & Koidis, A. (2022). Hyperspectral imaging
(HSI) for meat quality evaluation across the supply chain: Current and future
trends. Current Research in Food Science, 5, 1017-1027.
https://doi.org/10.1016/j.crfs.2022.05.016
JMGA (Japan Meat Grading Association). (2000). Beef Carcass
Grading Standards. Tokyo. Japan. Retrieved from
http://wagyu.org/uploads/page/JMGA%20Meat%20Grading%20Brochure_english.pdf
Khaled, A. Y., Parrish, C. A., & Adedeji, A. 2021. Emerging nondestructive approaches for
meat quality and safety evaluation—A review. Comprehensive Reviews in Food
Science and Food Safety, 20(4), 3438-3463.
https://doi.org/10.1111/1541-4337.12781
Khan,
A., Munir, M. T., Yu, W., & Young, B. (2020). Wavelength selection for
rapid identification of different particle size fractions of milk powder using
hyperspectral imaging. Sensors, 20(16), 4645.
https://doi.org/10.3390/s20164645
Li,
Y., Shan, J., Peng, Y., & Gao, X. (2011). Nondestructive assessment of beef-marbling
grade using hyperspectral imaging technology. In 2011 international conference
on new technology of agricultural (pp. 779-783). IEEE.
https://doi.org/10.1109/ICAE.2011.5943908
Liu,
J., Ellies-Oury, M. P., Stoyanchev, T., & Hocquette, J. F. (2022). Consumer
perception of beef quality and how to control, improve and predict it? Focus on
eating quality. Foods, 11(12), 1732.
https://doi.org/10.3390/foods11121732
Munera,
S., Amigo, J. M., Aleixos, N., Talens, P., Cubero, S., & Blasco, J. (2018).
Potential of VIS-NIR hyperspectral imaging and chemometric methods to identify
similar cultivars of nectarine. Food Control, 86, 1-10.
https://doi.org/10.1016/j.foodcont.2017.10.037
Nychas,
G. J., Sims, E., Tsakanikas, P., & Mohareb, F. (2021). Data Science in the
Food Industry. Annual Review of Biomedical Data Science, 4, 341-367.
https://doi.org/10.1146/annurev-biodatasci-020221-123602
Özdoğan,
G., Lin, X., & Sun, D. W. (2021). Rapid and noninvasive sensory analyses of
food products by hyperspectral imaging: Recent application developments. Trends
in Food Science & Technology, 111, 151-165.
https://doi.org/10.1016/j.tifs.2021.02.044
Pereira,
P. M. C. C., & Vicente, A. F. R. B. (2013). Meat nutritional
composition and nutritive role in the human diet. Meat science, 93(3),
586-592. https://doi.org/10.1016/j.meatsci.2012.09.018
Pinto,
D. L., Selli, A., Tulpan, D., Andrietta, L. T., Garbossa, P. L. M., Vander
Voort, G., ... & Ventura, R. V. (2023). Image feature extraction via local
binary patterns for marbling score classification in beef cattle using
tree-based algorithms. Livestock Science, 267, 105152.
https://doi.org/10.1016/j.livsci.2022.105152
Siche, R., Vejarano, R., Aredo, V., Velasquez, L., Saldana, E., & Quevedo, R. (2016). Evaluation of food quality and safety
with hyperspectral imaging (HSI). Food Engineering Reviews, 8(3),
306-322. https://doi.org/10.1007/s12393-015-9137-8
Su, W. H., & Sun, D. W. (2018). Multispectral
imaging for plant food quality analysis and visualization. Comprehensive
reviews in food science and food safety, 17(1), 220-239.
https://doi.org/10.1111/1541-4337.12317
Velásquez, L., Cruz-Tirado, J. P., Siche, R., & Quevedo, R. (2017). An application based on the decision
tree to classify the marbling of beef by hyperspectral imaging. Meat Science,
133, 43-50. https://doi.org/10.1016/j.meatsci.2017.06.002
Vidal,
P. O., Cardoso, R. D. C. V., Nunes, I. L., & Lima, W. K. D. S. (2022).
Quality and Safety of Fresh Beef in Retail: A Review. Journal of Food
Protection, 85(3), 435-447. https://doi.org/10.4315/JFP-21-294
Wold,
S., Sjöström, M., & Eriksson, L. (2001). PLS-regression: a basic tool of
chemometrics. Chemometrics and intelligent laboratory systems, 58(2),
109-130. https://doi.org/10.1016/S0169-7439(01)00155-1
Xie,
C., Chu, B., & He, Y. (2018). Prediction of banana color and firmness using
a novel wavelengths selection method of hyperspectral imaging. Food
chemistry, 245, 132-140. https://doi.org/10.1016/j.foodchem.2017.10.079
![]() https://orcid.org/0000-0002-3293-7696 L.
Velásquez:
https://orcid.org/0000-0002-3293-7696 L.
Velásquez: ![]() https://orcid.org/0000-0001-8091-5611
https://orcid.org/0000-0001-8091-5611![]() https://orcid.org/0000-0002-7174-8337
https://orcid.org/0000-0002-7174-8337